Optimization of Brain Penetrant 11Beta-Hydroxysteroid Dehydrogenase Type I Inhibitors and in Vivo Testing in Diet- Induced Obese Mice.
Goldberg, F.W., Dossetter, A.G., Scott, J.S., Robb, G.R., Boyd, S., Groombridge, S.D., Kemmitt, P.D., Sjogren, T., Gutierrez, P.M., Deschoolmeester, J., Swales, J.G., Turnbull, A.V., Wild, M.J.(2014) J Med Chem 57: 970
- PubMed: 24422550
- DOI: https://doi.org/10.1021/jm4016729
- Primary Citation of Related Structures:
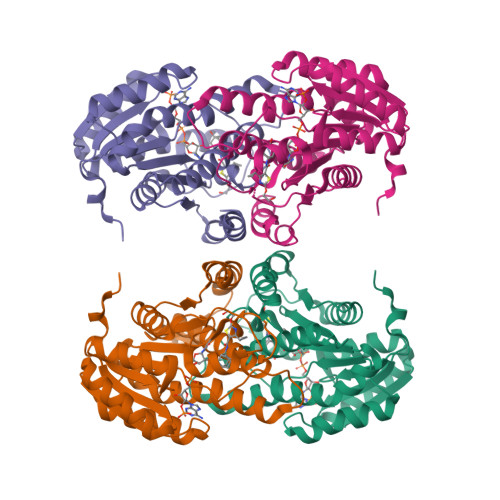
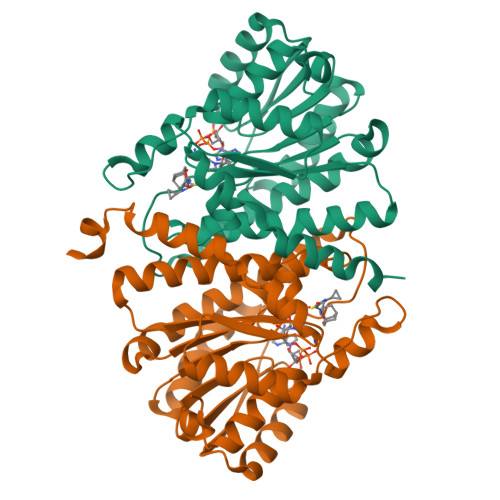
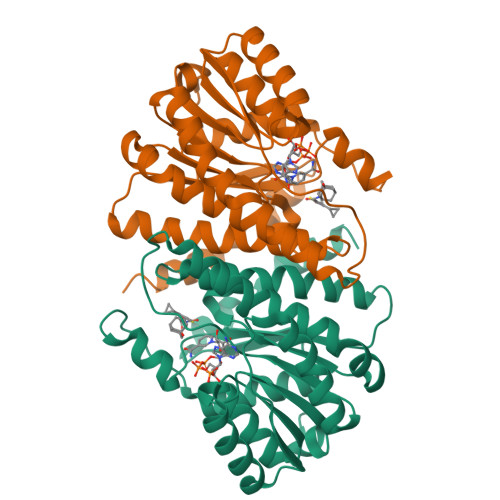
4C7J, 4C7K - PubMed Abstract:
11β-Hydroxysteroid dehydrogenase type 1 (11β-HSD1) has been widely considered by the pharmaceutical industry as a target to treat metabolic syndrome in type II diabetics. We hypothesized that central nervous system (CNS) penetration might be required to see efficacy. Starting from a previously reported pyrimidine compound, we removed hydrogen-bond donors to yield 3, which had modest CNS penetration. More significant progress was achieved by changing the core to give 40, which combines good potency and CNS penetration. Compound 40 was dosed to diet-induced obese (DIO) mice and gave excellent target engagement in the liver and high free exposures of drug, both peripherally and in the CNS. However, no body weight reduction or effects on glucose or insulin were observed in this model. Similar data were obtained with a structurally diverse thiazole compound 51. This work casts doubt on the hypothesis that localized tissue modulation of 11β-HSD1 activity alleviates metabolic syndrome.
Organizational Affiliation:
AstraZeneca , Mereside, Alderley Park, Macclesfield SK10 4TG, United Kingdom.